Digital PCR Device Materials
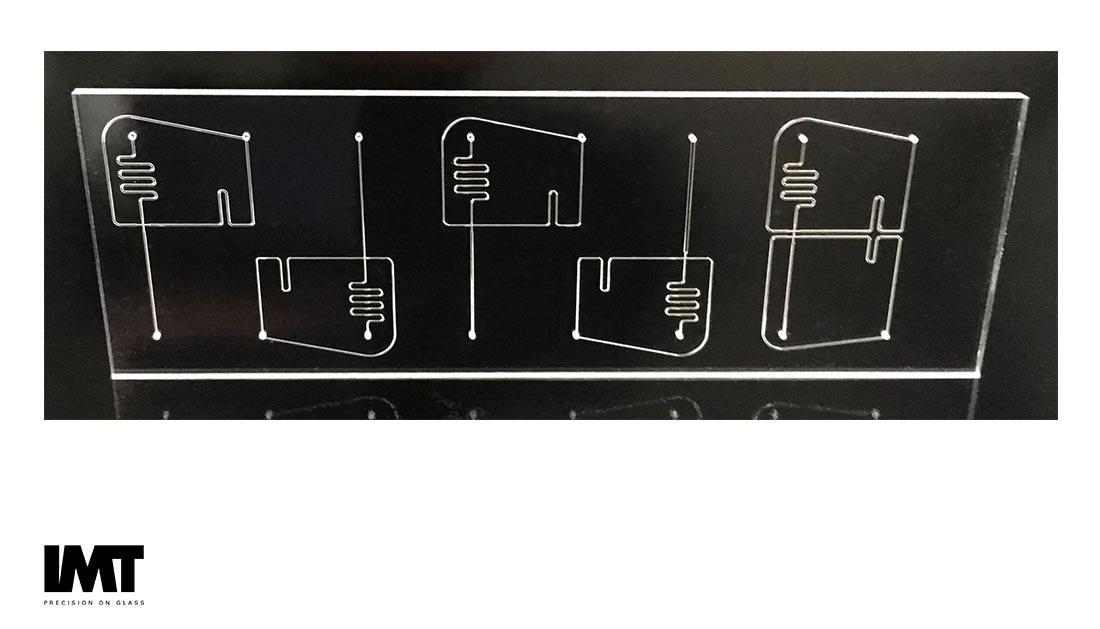
Many diagnostic approaches use the polymerase chain reaction (PCR), an enzymatic approach to sample pre-concentration, as an amplification step to reduce the amount of genetic material required for analysis. This is particularly effective with microfluidic approaches where the total device volume is quite small, meaning the actual concentration of DNA can become very high.
The best material for high-throughput, small volume nucleic acid analysis is largely determined by the detection mechanism and thermal properties of the materials. For optical detection methods, glass is still the material of choice. However, with electronic detection methods proliferating, other materials from silicon to polymers are suitable.
Glass is an attractive substrate for other reasons – fused silica does not have a strong inhibition effect on PCR enzymes [1]. It does not outgas or take on water. Most importantly, the glass surface does not store or release undesirable ions that can foul an enzyme reaction.
One of the major innovations achieved by the drive to reach the $1000 genome is the recognition of the role of sample preparation including the amplification and quantitation of DNA libraries using digital PCR. Digital PCR (dPCR)has been in existence for since 1992 by P. J. Sykes, et al. [2], and is a method of digitizing PCR by separating the sample into many partitions to try and get to a single molecule per partition. With smaller partitions, the ability to quantify different sequences whose presence occurs in small percentages (<1 %) increases. This was originally done using multiplexed capillaries and arrays in 384 well plates. Normal sample volumes for this array approach, however, required relatively large volumes and were dauntingly expensive [3].
Microfluidic devices designed to create droplets from an emulsion in a small volume version of flow injection analysis (droplet microfluidics) or harness electrowetting on an electrode array to create and move droplets (digital microfluidics), have come to dominate the approach to dPCR. Droplet digital and digital microfluidics has allowed for the confining of single molecules, single cells, and extremely small volumes of many reagents in a wide range of applications.
The ability to create thousands to millions of uniformly sized droplets is key to success for this approach, and this is heavily dependent on the design and manufacture of the digital microfluidic chip. The end result is better precision and selectivity when used for either standalone dPCR or dPCR that is a part of the NGS workflow. These advances have been applied to improve the understanding complex diseases and expression pathways such as cancer and is now being used in theranostics approaches such as liquid biopsies [4].
Companies such as Advanced Liquid Logic (now part Illumina), Raindance, and Quantalife (both now part of BioRad) have perfected creating droplets using the geometry and surface chemistry of the microfluidic channels as they interact with the droplet-enclosed fluid (hydrophilic) and droplet enclosing fluid (hydrophobic). The droplet digital PCR, or ddPCR, chip is designed to create droplets by combining an aqueous and hydrophobic liquid using pressure driven flow, hydrophilic and hydrophobic surface chemistry, and channel geometry [5]. Stilla Technologies’ product, the Naica System, combines the ddPCR (which Stilla calls droplet crystals) on a planar array format and performs quantitation on the same chip where PCR is performed using only 3 colors [6]. This considerably simplifies the workflow, reducing the need for reagents, which is important in making ddPCR in vitro analysis applications such as liquid biopsy easier to use.
Glass plays a major role in the successful manufacture of pressure-driven droplet microfluidic components since it provides excellent surface properties, e.g. low surface roughness, chemical inertness and high accuracy of manufacturing tolerances to ensure reproducible droplet volume. Additionally, because of the well-known surface chemistry, it is relatively easy to pattern hydrophobic regions on hydrophilic glass to create uniform droplet volumes and aid in droplet sorting.
Digital microfluidics uses an array of electrodes and electrowetting to create and manipulate droplets [7]. Electrowetting is the property of an applied voltage to modify the wettability of a surface. When an aqueous solution hits a hydrophobic electrode, surface forces cause the solution to bead up, forming a droplet. Applying an electric field between the droplet and the electrode allows the droplet to spread out. By creating an electrode pattern, it is possible to control droplet size and movement. For this effect, glass is the best material, as the high-density ITO electrodes required are readily patterned on glass.
With the $100 genome now within sight, it is more important than ever to make use of the advantages that come from combining microfabrication, microfluidics, and PCR. Working in continuously flowing streams, droplet microfluidic enables digital PCR in ever decreasing volumes. Digital microfluidics utilizes the ability to pattern electrodes in shrinking dimensions and control PCR reactions via electrowetting. The end result is a high-throughput sample prep that scales with NGS making sequencing faster, cheaper, and better.
Works Cited
[1] R. Kodzius, K. Xiao, J. Wu, X. Yi, X. Gong, I. G. Foulds and W. Wen, "Inhibitory effect of common microfluidic materials on PCR outcome," Sensors and Actuators B: Chemical, vol. 161, pp. 349-358, 2012.
[2] P. J. Sykes, S. J. Neoh, M. J. Brisco, E. Hughes, J. Condon and A. A. Morley, "Quantitation of targets for PCR by use of limiting dilution.," vol. 13, no. 3, pp. 444-449, 1992.
[3] M. Baker, "Digital PCR hits its stride," vol. 9, pp. 541-544, 2012.
[4] G. Karlin-Neumann, "http://www.americanlaboratory.com/914-Application-Notes/183421-Improved-Liquid-Biopsies-With-Combined-Digital-PCR-and-Next-Generation-Sequencing/," Amerlican Laboratory, 19 February 2016.
[5] S. Mashaghi, A. Abbaspourrad, D. A. Weitz and A. M. van Oijen, "Droplet microfluidics: A tool for biology, chemistry and nanotechnology," TrAC, vol. 82, pp. 118-125, 2016.
[6] Stilla Technologies, "Naica System," [Online]. Available: http://www.stillatechnologies.com/naica-system/.
[7] K. Choi, A. H. Ng, R. Fobel and A. Wheeler, "Digital Microfluidics," Annu. Rev. Anal. Chem. , vol. 5, pp. 413-440, 2012.